RESEARCH INTERESTS
Social organisms, from microbes to humans, rank among the most abundant and ecologically dominant species on Earth. These interspecies interactions rely on complex molecular, chemical and behavioral signals that are the products of underlying evolutionary changes in their genomes. Symbiotic associations also require coevolution between partners to reinforce their relationship. At the core of our work, we seek to understand the molecular and cellular phenomena that underpin symbiotic living, and how these lifestyles in turn impact the evolution of symbiotic species’ genomes.
The lab focuses on studying marine symbioses, how climate change affects these associations, and applied solutions for their conservation. We seek to identify the degree to which symbiotic partners can acclimate and/or adapt to changing environments. By employing quantitative and empirical approaches to both field-based and laboratory model systems, we work on the following research themes.
The lab focuses on studying marine symbioses, how climate change affects these associations, and applied solutions for their conservation. We seek to identify the degree to which symbiotic partners can acclimate and/or adapt to changing environments. By employing quantitative and empirical approaches to both field-based and laboratory model systems, we work on the following research themes.
Partner specificity and fidelity
|
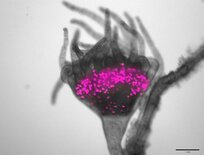
The sympatric corals Acropora palmata and A. cervicornis form the hybrid, A. prolifera. Symbionts of these corals (Symbiodinium 'fitti') are structured by their host taxa—not geographic location—suggesting ecological specialization to a hybrid host. The symbiont generation time inside a coral host can range from 3-75 days, or about 14.6-365 gens/host gen. Thus, hybrid corals provide novel genetic backgrounds for symbiont interactions and evolution. We use the Acropora-S. 'fitti' association to study how evolutionary, geographic, ecological, and abiotic factors shape symbiont interactions in novel hosts.
|
Interspecies communication under environmental stress
|
Symbiosis-driven lipid signaling and the cnidarian heat stress response are evolutionarily conserved processes that remain largely unexplored phenomena at the molecular level. Our lab investigates molecular mechanisms underlying these conserved pathways in a variety of model systems, including Exaiptasia and the symbiotic jellyfish Cassiopea —two cnidarians that partner with similar dinoflagellate symbionts as corals. We use functional tools like RNAi, CRISPR and pharmacological agents with controlled stress experiments in the lab.
|
Genomic patterns of convergence in mutualistic symbioses
|
Mutualistic associations between various organisms and photosynthetic, unicellular algae are found repeatedly across the tree of life. Such organisms represent natural, replicated evolutionary experiments that can be probed to uncover analogous molecular features driving their phenotypic similarity. Large benthic foraminifera and sea anemones can partner with diverse photosynthetic groups such as dinoflagellates, chlorophytes, rhodophytes and diatoms. Thus, these convergent systems allow us to ask what genomic changes occur when lineages become symbiotic, and what subsequent changes occur for each algal type. We are using single-cell transcriptomes of non-symbiotic and symbiotic lineages to detect convergent changes. Identification of shared molecular adaptations in specific genes and pathways underlying symbiosis could reveal generalizable transitions toward mutualism with algae.
|