'Omic Resources
Chromosome-level Genome Assembly of Rove Beetle Dalotia
 Photo: Joe Parker Photo: Joe Parker
We've assembled the Dalotia genome using a hybrid approach with Oxford Nanopore and Illumina data. The scaffolds were ordered and elongated using a chromosomal interaction method called SPRITE . Premliminary assembly has 10 pseudomolecules with N50 of 13 Mbp and contains 98% of the assembled genome. This is an ongoing project with collaboration between the Parker lab and the Guttman lab a Caltech.
|

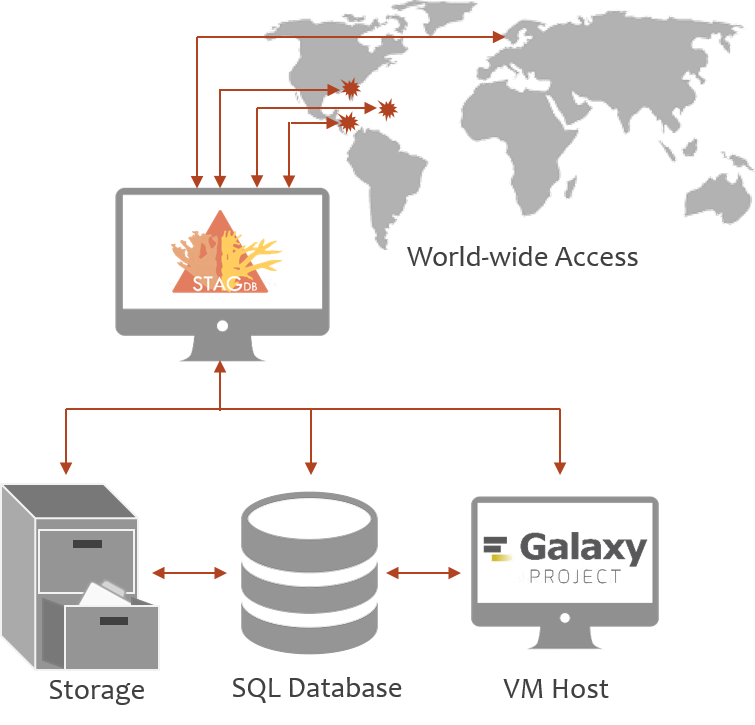
STAG: Standard Tools for Acroporid Genotyping
|
Current genotyping methods are restricted to a few labs creating a significant bottleneck to conservation projects. Recent identification of single nucleotide variants (SNVs) between species and populations allows for standardized SNV genotyping at comparable cost. We developed SNV probes and an automated analysis pipeline for multi-locus genotyping to inform management recommendations and standing genetic diversity of acroporids and their symbionts.
Publication on the SNPchip, STAG workflow and database available now. Code and database schema available on github. |
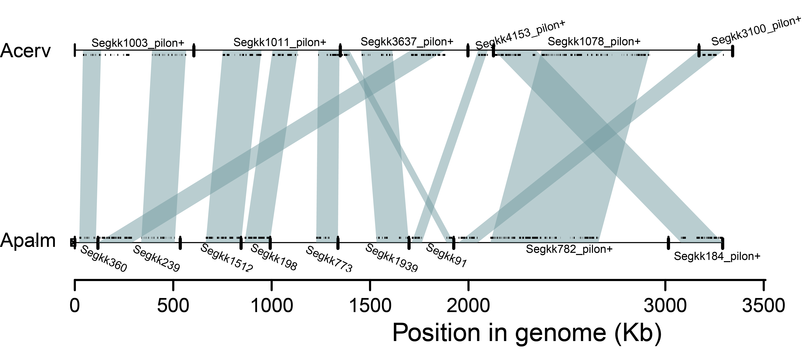
Caribbean Acroporid Genomes
|
Draft genome assemblies available here!
Construction of de novo genome assemblies of the Caribbean Acroporids, A. palmata and A. cervicornis are underway. These resources will allow us to explore patterns of introgression between these species and their hybrid, A. prolifera. (See associated work on Coral Hybridization) The data provided through this project constitutes the collaborative work of the myself and the Baums's Lab at Pennsylvania State University, Webb Miller at Pennsylvania State University and Fogarty's Lab at UNC Wilmington. |
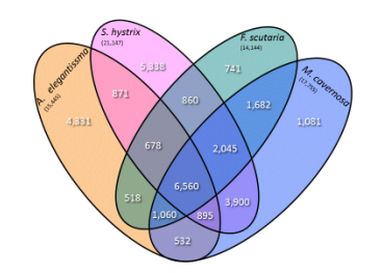
Anthozoan Transcriptomes
|
Through a collaboration with Dr. Eli Meyer, the Weis Lab sequenced the transcriptomes of the anemone A. elegantissma, and corals F. scutaria, M. cavernosa and S. hystrix. In the analysis of these transcriptomes, we constructed one of the first cnidarian phylogenomic trees.
See: Kitchen et al. 2015, G3 Transcriptomes available at reefgenomics.org |